Solutions

-
Overview
Some of the services and solutions we at New Atlantic Technology Group, LLC have developed over the years are now offered to the wider community. They are tools and solutions for diverse IT problems we have been working on. Generally, the research-related tools from the list below are open-source solutions and are part of our long term commitment to disseminate these utilities to the biomedical IT community as part of our collaboration with The National Center for Biomedical Computing Informatics for Integrating Biology and the Bedside, i2b2.
-
Tools for the genomics-enabled Translational Research Enterprise
-
Correlation Analysis Cell, an Eclipse plugin part of the i2b2 hive
New Atlantic Technology Group, LLC has released the Correlation Analysis plug-in, a major specialty-analysis Cell from the i2b2 hive. The i2b2 hive is the scalable computational platform the National Center for Biomedical Computing Informatics for Integrating Biology and the Bedside (i2b2) has been setout to develop and disseminate.
The Correlation Cell was developed by NATG, LLC and i2b2 under a contract supported by a grant from Partners HealthCare System, Inc. It is an open-source effort whose distribution is governed by the i2b2 Software License Agreement ("Software License") (see it here).
NATG, LLC will provide additional support options for the adopters and the users of the Correlation Analysis Cell. It will assume full second-level support of the application including bug fixes and enhancements. Please review our Services page for the specific support, implementation training and interpretaion service options we offer to i2b2 adoptees and other interested parties.
Please contact us with a description of the problem you are experiencing. We will contact you back within 4 hours during regular business hours or the next business day. You could also check this blog dedicated to Relevance Networks computation and implementation issues for similar posts or to submit a post.
Local Mirror for the i2b2's Correlation Cell, ver 1.0:Source for the Correlation Analysis Cell, requires Java 1.5 or better Installation Guide User's Guide Developer's Guide Tutorial Cell Messaging Guide
-
Standalone RelNet Tool
The standalone RelNet Tool is an open-source java based application that exposes a large part of the Correlation Analysis Cell functionality (see above) and doesn't require installation of the basic i2b2 hive. Consequently, it can work with a very broad range of data types, not only with the ones supported by the Hive concept ontologies.
The RelNet application is intended to complement an early, currently unsupported version of the Relevance Networks software developed at the Boston Children Hospital's Informatics Program (CHIP). Please refer to the CHIP site for more information regarding the publications and credits information.
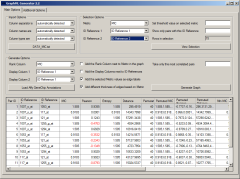
The list of the major functionality provided by the standalone RelNet Tool is as follows: Construct correlation vectors from a wide set of medical, genomic, proteomics and other biological data Visual interface and tools to split large GEO (NCBI's Gene Expression Omnibus) data files, sort by type of expression data format and import data On the fly computation of correlation profiles and ability to analyze results using a visual querying interface Construct visual representation of RelNets in the GraphML format using the free viewer from yWorks, yEd For expression data: visual interface for import of gene annotation and lookup tables, for various Affymetrix gene chips Ability to save results to a persistent storage (a database engine) for later review and or comparison with other correlation computation runs A comprehensive help system with information about setup, computation, import and analysis of correlation analysis results including visual representation of RelNets as GraphML formatted networks
Current Version: ver 2.1. Unzip and run RelNet.jar. Requires Java 1.6 or better.
Older versions: ver 2.0, ver 1.0.Please contact us if you need the complete source code of the RelNet standalone tool, or with a description of the problem you are experiencing.
-
GraphML Generator for RelNets
The GraphML Generator is a visual tool that greatly facilitates creation of a full-functionality GraphML files from flat files with correlation results. It can be used to quickly construct the visual representation of the multiply-connected graphs that are often the output of integrative studies in the modern genomic medicine. The Generator allows also for an advanced array of compounding and visual elements properties control. The GraphML files that the Generator creates are then displayed using the free viewer from yWorks, yEd.
The list of the the major functionality provided by the Generator is as follows: Visual interface for import of flat files with standard formatting of list of correlation pairs Ability to define subsets of the original result set based on various criteria as pair_id, threshold, number of pairs, presence/absence of given element in the pair etc. Define and apply visual representation styles on various levels: single node, groups of nodes Define and apply node annotation styles via import of external annotation lookup files The GraphML Generator is a Microsoft .NET 2.0+ based desktop application for Windows The source code is available upon request
Current Version: ver 2.2. Unzip and run GraphMLGenerator.exe. Requires .NET 2.0 or better.
Older versions: ver 2.1, ver 2.0, ver 1.0.Please contact us if you need the complete source code of the GraphML Generator, or with a description of the problem you are experiencing.
-
Expanding i2b2 to support Appropriateness research in Radiology: Number Of Studies Query capability and MID Analytics Client Plugin
New Atlantic Technology Group has developed the new core i2b2 functionality in close collaboration with the Center for Evidence Based Imaging (CEBI).
We have implemented new type of search in i2b2. Before data and application models of i2b2 resolve around its fundamental functionality of patient cohort discovery. Current queiries in i2b2 return either patient counts or patient lists. We developed a new search which returns number of studies. It allows the adoption of the i2b2 platform in primary HIT settings whose goal is to create administrative portals and executive dashboards, or in dual-purpose environments supporting the traditional patient cohort discovery mode, together with operations and utilization management research studies.
In addition, we have implemented an i2b2 instance with the number elf studies functionality and the novel MID Analytics Plugin Client.
Driving Project Background
Through the Medicare program, the Centers for Medicare & Medicaid Services (CMS) provide health insurance coverage to approximately 47 million Americans. Because of the Medicare program's large scale, changes in Medicare have the potential to affect a large number of beneficiaries and providers and often affect the overall health system. Therefore, when introducing innovations in the Medicare program or making significant changes in programs or policy, CMS often implements demonstration projects to test the impact of the changes on a smaller scale.
The Medicare Imaging Demonstration (MID) was authorized by the US Congress in the Medicare Improvements for Patients and Providers Act of 2008. The goal of the demonstration is to assess whether the use of decision support systems (DSS) that promote appropriate use of imaging services based on medical specialty guidelines, can improve quality of care and reduce unnecessary radiation exposure.
Through the MID project, CMS collects data on physician compliance with appropriateness criteria for imaging services. The demonstration will examine the impact of using a DSS on physicians' rate of ordering advanced imaging services and the appropriateness of the orders. Existing coverage and payment policies under Medicare do not change under this demonstration. The MID is focused on three advanced imaging modalities: magnetic resonance imaging (MRI), computed tomography (CT) and nuclear medicine. Within those modalities, the demonstration targets 11 of the most frequently used advanced imaging procedures.
Data Mapping Architecture
The mapping of the CMS-mandated data to the i2b2's Star Schema utilizes code modifiers to account for the multiplicity of the atomic observations recorded during the interaction of the ordering physician with the DSS system. For example, each of the distinctive steps of data collection during the ordering process (initial exam to be ordered based on signs/symptoms and differential diagnoses, characterization of the disease state of the patient via up to 10 diagnoses using their ICD9s, the alternative imaging studies suggested a) by the system and b) by the radiologist, and the final exam performed) results in multiple rows in the Observation_Fact table. Additional imaging exam types, CPT4, appropriateness and utilization management ontologies were built to facilitate store and querying of the data.
The new core i2b2 functionality was developed by the Center for Evidence Based Imaging (CEBI), a Center of Excellence of the Brigham and Women's Hospital in Boston, Massachusetts, in close collaboration with New Atlantic Tech Group, LLC of Newton, Massachusetts.
Version 1.6.09 of the i2b2 core server was used for implementing the #studies query. Download the complete archive here and make the server following the corresponding Install Guide. The changes in the sources and the database are detailed here.
-
Solutions for Telemedicine
New Atlantic Technology Group, LLC has been involved in developing new and integrating existing applications and utilities across the back-and-front-office environment of the modern filmless and paperless radiology department.
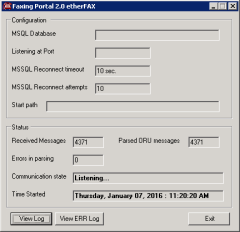
One such telemedicine solution allows for a seamless, automated and highly customizable delivery of radiology interpretation results back to their original ordering sites. The general functionality features of the Results Delivery Portal are as follows: integration with HL7-based direct, real-time HIS or PACS feed for a high-volume interpretation results scheduling and delivery rules-based engine for defining and customizing multiple delivery sites based on various fields in the HL7 header interface with third-party commercial automated faxing delivery interfaces a high degree of customizability of site delivery profiles including rules-based logic for multiple delivery attempts visual interface for easy, one-click manual resend complete back-log of all results communication messages, searchable by various parameters list of canned reports regarding status and delivery information of results, searchable by various parameters one-click identification of problematic fax messages, with display of a complete status information and ability to manually resend robust technological implementation as a multi-tier .NET 2.0+ application: a high-volume HL7 listener with a parsing engine, communication engine and web-based reports application for easy viewing and manipulation
Please contact us if you would like to learn more how our expertise in this area of telemedicine can serve you to address your particular needs.
-
Notification Solutions for Radiologists
Measuring and improving the overall radiology reports generation and finalization ('signing') time is becoming one of the important quality characteristics of the modern radiology enterprise. The truly electronic (filmless and paperless) radiology environment often relies on technologiest like Computerized Physician Order Entry (CPOE), digital Picture Archiving and Communication Systems (PACS) and various digital dictation and voice-recognition components for a delivery of timely and error-free diagnostic imaging services.
NATG, LLC has been instrumental in developing, implementing and supporting solutions for enabling measurement and notification of members of the radiology staff about their turn-around times.
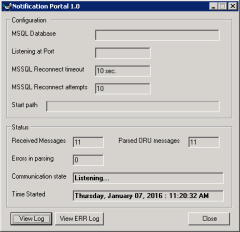
The general functionality features of the Notification for Radiologists Portal are as follows: integration with HL7-based direct, real-time RIS/PACS/Broker feed of all studies with the appropriate RIS status rules-based engine for scheduling and delivering of notification pages with number of unsigned reports interface with third-party alpha-numeric paging service for delivery of notification messages highly flexible and customizable rules-based interface for defining how/when to notify radiologists ability to elevate status of notification to supervisors interface to schedule application status notification via email/pager to admin staff analytical reports to display average turn-around time per radiologist and level of conformation to departmentally accepted standards one-click identification of problematic fax messages, with display of a complete status information and ability to manually resend robust technological implementation as a multi-tier .NET 2.0+ application: a high-volume HL7 server with a parsing engine, Messaging engine and web-based reports application
Instances of the Results Delivery and the Status Notification portals have been customized and used in routine clinical care for over 5 years now, in the highly complex IT environment of a large Northeast IDN-based hospital, in downtown Boston.
Please contact us if you would like to learn more about this solution.